CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is an acronym for DNA loci that contain multiple, short, direct repetitions of base sequences. Each repetition contains a series of base pairs followed by the same or a similar series in reverse and then by 30 or so base pairs known as "spacer DNA". CRISPRs are found in the genomes of approximately 40% of sequenced bacteria and 90% of sequenced archaea. CRISPRs are often associated with cas genes which code for proteins that perform various functions related to CRISPRs. The CRISPR-Cas system functions as a prokaryotic immune system, in that it confers resistance to exogenous genetic elements such as plasmids and phages and provides a form of acquired immunity. CRISPR spacers recognize and silence exogenous genetic elements in a manner analogous to RNAi in eukaryotic organisms.
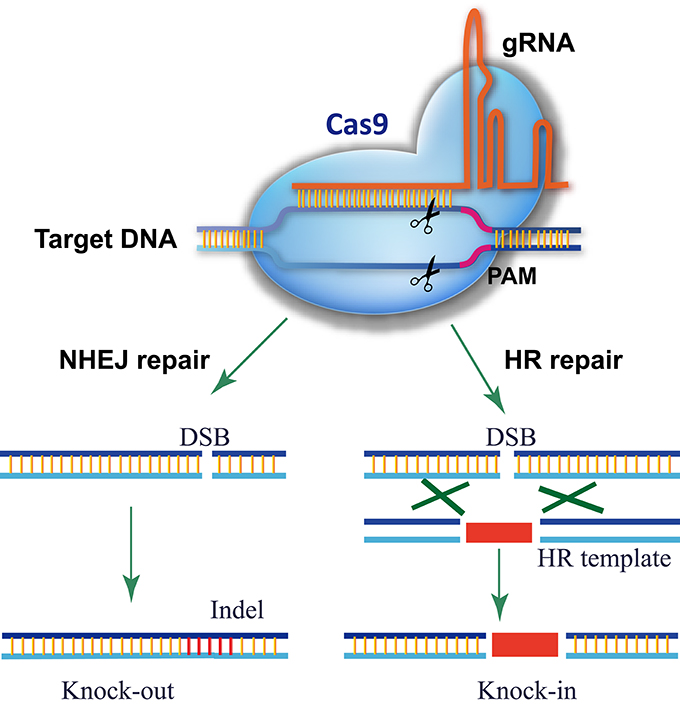
CRISPR-P 2.0 design tool based on we previously work of CRISPR-P (Lei et al., 2014), which is one of the most popular tools for sgRNA design in plants. As CRISPR/Cas9 technology has evolved rapidly in the past two years, an updated platform, CRISPR-P 2.0 provides web service for computer-aided design of highly efficient sgRNA with minimal off-target effects. It has functions as follow:
- It supports sgRNA design for 49 plant genomes. SgRNAs for 49 plant species with the latest version of genome and annotation are provided.
- Scoring system of sgRNA for on-target efficiency and off-target potential are improved. CRISPR-P 2.0 uses a modified scoring system to rate the off-targeting potential and on-targeting efficiency of sgRNAs for Streptococcus pyogenes Cas9 which is the widest used CRISPR-Cas9 system. The scoring system in CRISPR-P 2.0 is based on the up-to-date knowledge about SpCas9 genome editing.
- It Supports various CRISPR-Cas systems. It supports to design guide sequences for various CRISPR-Cas systems like Cpf1 (Zetsche et al., 2015) and various Cas9 endonucleases.
- A comprehensive analysis of the guide sequence is provided, including: GC content, restriction endonuclease site, microhomology sequence flanking the targeting site (microhomology score), and the secondary structure of sgRNA.
- Identification of sgRNA from custom sequences is also provided. If user’s genome/sequence is not listed in the selectable genomes, it allows users to upload custom sequences and identify sgRNAs.
Visit the old version of CRISPR-P
Reference
1. Yang Lei, Li Lu, Hai-Yang Liu, Sen L, Feng Xing, Ling-Ling Chen*. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant, 2014, 7(9): 1494-1496. doi: 10.1093/mp/ssu044.
2. Yuduan Ding, Hong Li, Ling-Ling Chen*, Kabin Xie*. Recent advances in genome editing using CRISPR/Cas9. Front Plant Sci., 2016, 7: 703.PMC4877526.
3. Hao Liu, Yuduan Ding, Yanqing Zhou, Wenqi Jin, Kabin Xie*, Ling-Ling Chen*. CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants. Mol Plant, 2017, 10(3): 530-532. doi: 10.1016/j.molp.2017.01.003 .
